Research on human skin diseases has been hampered by limitations on the number of studies that can be done on formalin-fixed, paraffin-embedded (FFPE) specimens, the most frequent type of specimens that are generated by clinical care of dermatology patients, research studies and clinical trials.
This core makes three transformative techniques, tyramide based immunostaining combined with spectral imaging, NanoString based gene expression profiling, and cutting edge digital spatial profiling, available to any skin disease researcher at any institution to allow human skin disease researchers to derive comprehensive phenotypic, functional and gene expression analyses, all from a single small biopsy of skin.
Tyramide Signal Amplification (TSA) Staining & Multispectral Imaging
Serial TSA staining is a simple, elegant way to sequentially immunostain histologic sections for six different markers without interspecies interference. It can be used to quantify even low abundance proteins. Slides are imaged on the Center’s Akoya Biosciences Mantra Quantitative Pathology Station microscope system which allows separation of up to 6 markers into separate channels. Image analysis with the integrated inForm software allows automated cell identification and phenotyping.
The Center provides access to validated TSA antibody staining panels, staining protocols, access to the Mantra imaging microscope to image samples and access to the inForm software for automated cell analyses.
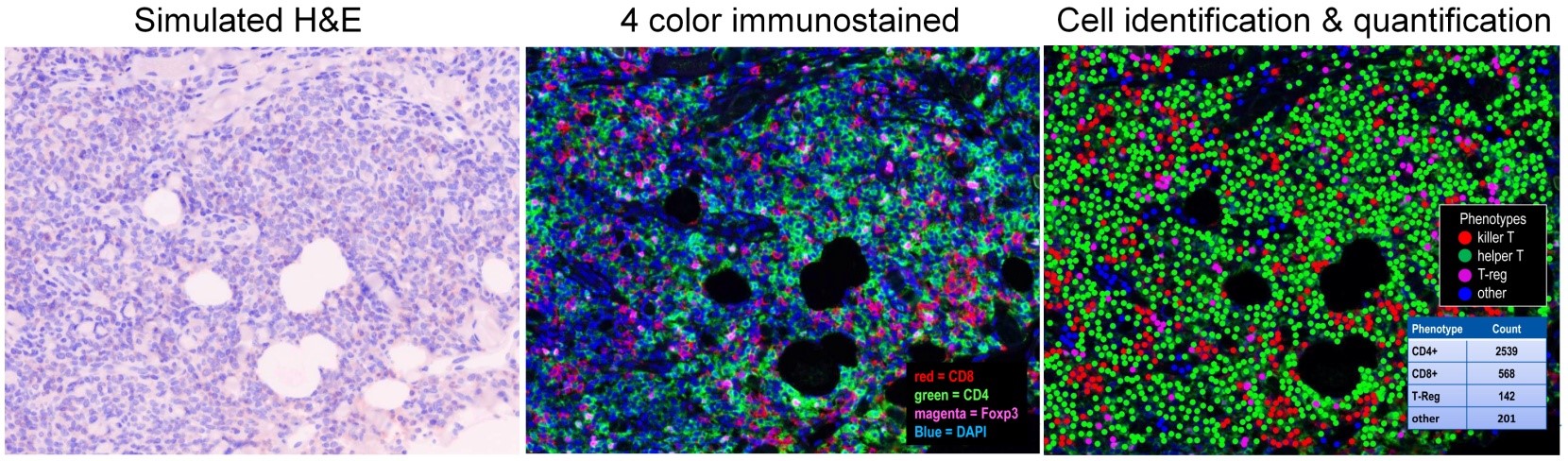
A skin lesion from a patient with stage IA cutaneous T-cell lymphoma was serially stained with CD4, CD8 and FOXP3 specific monoclonal antibodies and with DAPI (a nuclear stain). The immunostained sample (center) and the automated cell identification and counting analysis done on this stained section are shown (right). The total number of CD4+ T cells, CD8+ T cells and regulatory T cells in the section are shown in the table on the bottom right of the analyzed section.
NanoString based gene expression profiling
FFPE sample preservation maintains morphology but the cross-linking and degradation of RNA and DNA that occurs in FFPE samples has made it difficult to analyze nucleic acids in these tissues. NanoString’s probe-based nucleic acid detection technology has overcome these barriers, allowing quantification of up to 800 DNA, RNA, and miRNA targets from even highly degraded FFPE samples.
The Center provides assistance with experimental planning, RNA isolation, access to the n-Counter instruments needed to analyze samples, and custom panel design. The Center will also provide NanoString data from analysis of healthy human skin to help you gauge the significance of your results.
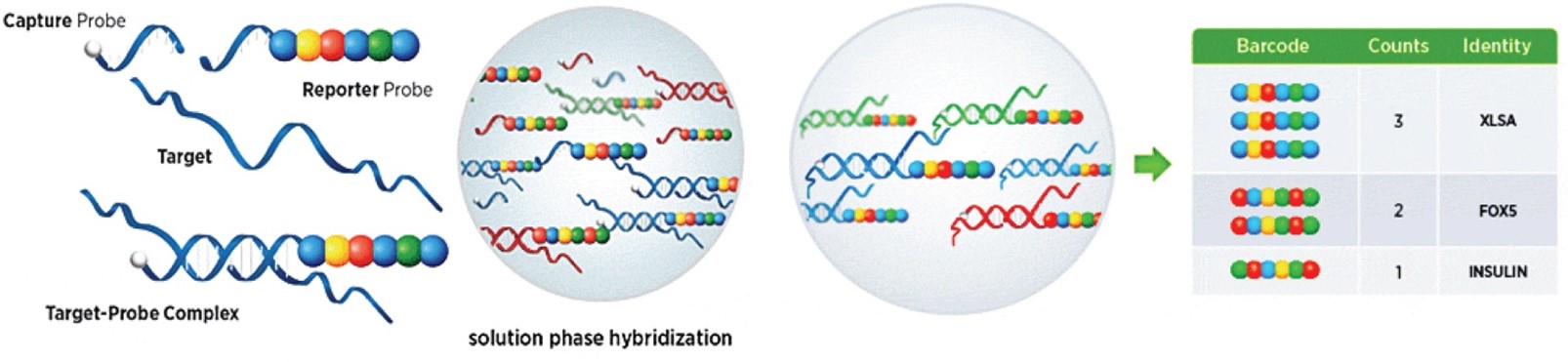
How it works: The reporter probe and capture probes are added to the nucleic acid suspension in solution, where they bind on a one-to-one basis with the specific DNA or RNA target. The biotin moiety on the capture probe is then used to immobilize the complex, and unbound probe is rinsed away. The complexes are then read using an n-Counter instrument. Image courtesy of NanoString.
Digital Spatial Profiling: The NanoString CosMx Spatial Molecular Imager
The CosMx Spatial Molecular Imager utilizes NanoString’s unique probe sets to directly measure mRNA levels for 1,013 standard genes, 50 customizable add in genes, expression levels of 100 proteins and spatial information at a subcellular resolution (<100 nm) on FFPE tissue sections. This approach generates true single cell gene expression, protein expression and spatial information about the proximity of each cell type to others.
The Center provides assistance with experimental planning and custom panel design as well as access to the instrument.
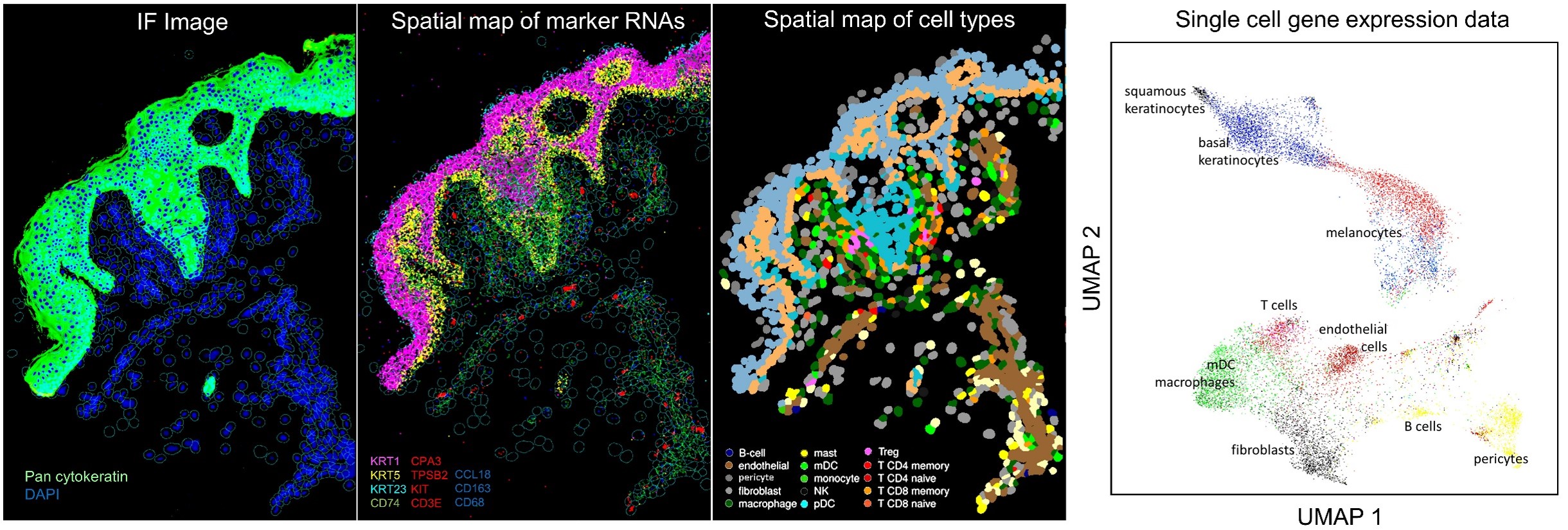
A FFPE section from a face transplant rejection biopsy was stained with pan-cytokeratin to delineate the epidermis (green, first panel) vs. dermis. Expression of cell type marker RNAs (second panel) was used to spatially assign cell types (third panel). These analyses produced true single cell data, with clear delineation of individual immune and stromal cell types (last panel).