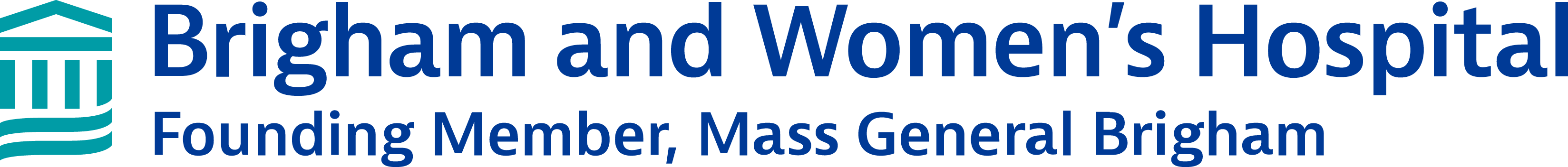Research into healthy human skin biology and skin diseases has been hampered by the lack of transgenic mouse models and the limited studies that can be performed in human beings. However, cutting-edge approaches have become available that are increasingly providing key mechanistic insights into human immunobiology.
This core makes three groundbreaking techniques – single cell RNA sequencing, cytometry by time of flight (CyTOF), and next-generation high throughput T cell receptor sequencing – available to any skin disease researcher in any geographic location, affiliated with any institution, to accelerate cutting edge research in human skin biology and skin diseases.
Single cell RNA sequencing
Molecular studies on whole tissues and enriched cell types provide a global picture of pathological processes, while single cell analyses provide a new look at the nature of cell types, cell states, and the molecular pathways that may be obscured in bulk populations. The Center makes available a variety of 10X Genomics technologies for single cell profiling with an integrated platform that includes technical expertise in processing, storing, and analyzing samples.
The Center offers access to the following single cell approaches on the 10X Genomics platform:
- Single cell RNA sequencing (scRNA-seq) – Capture of gene expression by individual cells from a viable sample
- Cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) – Addition of oligonucleotide-labeled antibodies into scRNA-seq analyses to simultaneously identify protein markers and RNA expression
- Sample batching – Combining multiple samples into a single run to allow direct comparisons between samples without confounding batch effects
- T cell receptor gene capture – Analysis of the rearranged alpha and beta chains of the T cell receptor in individual cells to study gene expression by individual T cell clones
- Single cell ATAC-seq – ATAC-seq identifies open chromatin sites and reveals the interplay between genomic locations of open chromatin, DNA binding proteins, individual nucleosomes, and chromatin compaction
The Center also offers enabling technologies and support for single cell analyses in the form of optimized skin preservation, shipping and digestion protocols, isolation and preparation of viable cells for single cell sequencing, and experimental planning assistance.
Single cell RNA sequencing results from skin biopsies sent to the Center. Skin biopsies from cutaneous T cell lymphoma were obtained before and after treatment with a novel therapy. Biopsies were sent to the Center and cryopreserved until the day of analysis. Samples were digested, flow sorted for viable CD45 + immune cells, batched and analyzed by scRNA-seq with TCR gene capture. Malignant T cells were identified by their expression of a clonal TCR. CITE-seq antibodies were included to identify tumor-specific T cells via their expression of CD39 and PD-1. CD8 T cells in the skin were increased after therapy and malignant T cells declined, indicating efficacy of the test drug.
Cytometry by Time of Flight (CyTOF)
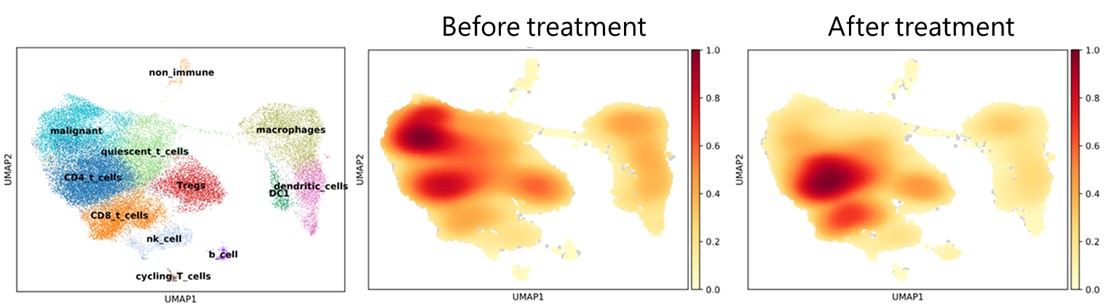
Flow cytometry is a powerful tool but the number of different markers that can be immunostained and detected on a particular cell is limited by spectral overlap of the fluorochromes used. Cytometry by time of flight (CyTOF) combines immunostaining with mass cytometry. Cells are immunostained with antibodies conjugated to rare earth metals and then analyzed by a mass cytometer, allowing highly quantitative measurement of 45 separate markers, with expansion to 75 labels in the future, with no signal overlap and no need for compensation.
The Center provides access to validated CyTOF antibody panels, staining protocols, access to CyTOF instruments to run samples and access to analysis techniques.
Advantages of CyTOF. Conventional flow cytometry (left) utilizes fluorophores that have overlapping spectra. As a result, there is an upper limit to the number of markers that can be studied at one time and increasingly complex compensation is required to separate the signals from different channels. In contrast, CyTOF (right) utilizes antibodies conjugated to rare earth metals. This allows measurement of up to 45 differently labeled antibodies and detection reagents, with expansion to 75 labels in the future, with no overlap and no need for compensation.
High Throughput T Cell Receptor CDR3 sequencing (HTS)
HTS is a remarkable technique that allows measurement of the absolute number of T cells, the diversity of the T cell repertoire as assessed by the number of different TCR sequences, the relative proportion of a particular T cell sequence in the total population and the exact CDR3 sequences of all T cells present in a particular biologic sample in a single, one step test. This test requires only 150 ng of DNA that can be extracted from blood (including flow sorted purified T cell populations) and both fresh and FFPE tissues specimens.
The Center provides assistance with experimental design (can your experiment answer the question?), isolation and sorting of cells from human skin, DNA isolation, shipping of samples to Adaptive Biotechnologies (where HTS is carried out) and comprehensive support in the analysis of HTS data.
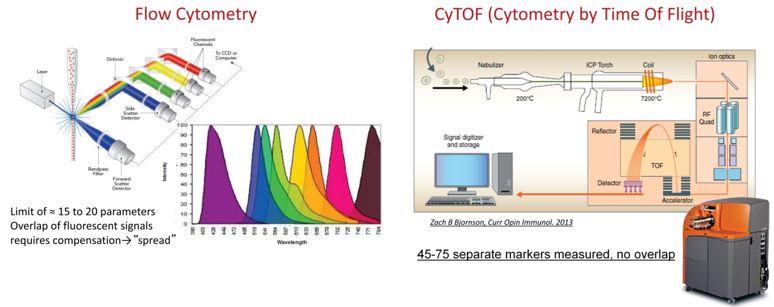
HTS of clinically resolved lesions following clearance on etanercept therapy demonstrated the presence of expanded residual T cell clones. HTS analyses were performed on the same psoriatic lesion before (left) and after) clearance on etanercept therapy.